Multinomial Logistic Regression
Prediction + model selection + conditions
Prof. Maria Tackett
Topics
Predictions
Model selection
Checking conditions
NHANES Data
National Health and Nutrition Examination Survey is conducted by the National Center for Health Statistics (NCHS)
The goal is to "assess the health and nutritional status of adults and children in the United States"
This survey includes an interview and a physical examination
Health Rating vs. Age & Physical Activity
Question: Can we use a person's age and whether they do regular physical activity to predict their self-reported health rating?
We will analyze the following variables:
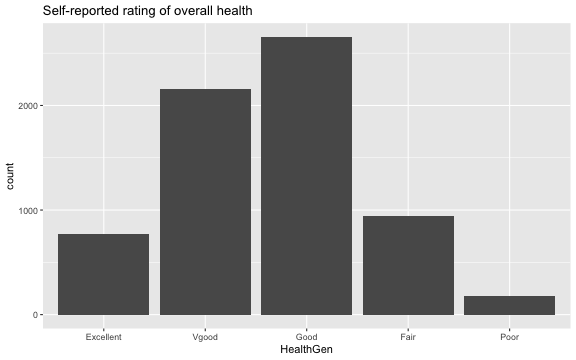
HealthGen: Self-reported rating of participant's health in general. Excellent, Vgood, Good, Fair, or Poor.Age: Age at time of screening (in years). Participants 80 or older were recorded as 80.PhysActive: Participant does moderate to vigorous-intensity sports, fitness or recreational activities
Model in R
| y.level | term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|---|
| Vgood | (Intercept) | 1.205 | 0.145 | 8.325 | 0.000 |
| Vgood | Age | 0.001 | 0.002 | 0.369 | 0.712 |
| Vgood | PhysActiveYes | -0.321 | 0.093 | -3.454 | 0.001 |
| Good | (Intercept) | 1.948 | 0.141 | 13.844 | 0.000 |
| Good | Age | -0.002 | 0.002 | -0.977 | 0.329 |
| Good | PhysActiveYes | -1.001 | 0.090 | -11.120 | 0.000 |
| Fair | (Intercept) | 0.915 | 0.164 | 5.566 | 0.000 |
| Fair | Age | 0.003 | 0.003 | 1.058 | 0.290 |
| Fair | PhysActiveYes | -1.645 | 0.107 | -15.319 | 0.000 |
| Poor | (Intercept) | -1.521 | 0.290 | -5.238 | 0.000 |
| Poor | Age | 0.022 | 0.005 | 4.522 | 0.000 |
| Poor | PhysActiveYes | -2.656 | 0.236 | -11.275 | 0.000 |
Predictions
Calculating probabilities
For categories 2,…,K, the probability that the ith observation is in the jth category is
ˆπij=exp{ˆβ0j+ˆβ1jxi1+⋯+ˆβpjxip}1+K∑k=2exp{ˆβ0k+ˆβ1kxi1+…ˆβpkxip}
For the baseline category, k=1, we calculate the probability ˆπi1 as ˆπi1=1−K∑k=2ˆπik
NHANES: Predicted probabilities
#calculate predicted probabilitiespred_probs <- as_tibble(predict(health_m, type = "probs")) %>% mutate(obs_num = 1:n())## # A tibble: 5 x 6## Excellent Vgood Good Fair Poor obs_num## <dbl> <dbl> <dbl> <dbl> <dbl> <int>## 1 0.0705 0.244 0.451 0.198 0.0366 101## 2 0.0702 0.244 0.441 0.202 0.0426 102## 3 0.0696 0.244 0.427 0.206 0.0527 103## 4 0.0696 0.244 0.427 0.206 0.0527 104## 5 0.155 0.393 0.359 0.0861 0.00662 105Add predictions to original data
health_m_aug <- inner_join(nhanes_adult, pred_probs, by = "obs_num") %>% select(obs_num, everything())## Rows: 6,710## Columns: 10## $ obs_num <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, …## $ HealthGen <fct> Good, Good, Good, Good, Vgood, Vgood, Vgood, Vgood, Vgood, …## $ Age <int> 34, 34, 34, 49, 45, 45, 45, 66, 58, 54, 50, 33, 60, 56, 56,…## $ PhysActive <fct> No, No, No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, …## $ Education <fct> High School, High School, High School, Some College, Colleg…## $ Excellent <dbl> 0.07069715, 0.07069715, 0.07069715, 0.07003173, 0.15547075,…## $ Vgood <dbl> 0.2433979, 0.2433979, 0.2433979, 0.2444214, 0.3922335, 0.39…## $ Good <dbl> 0.4573727, 0.4573727, 0.4573727, 0.4372533, 0.3599639, 0.35…## $ Fair <dbl> 0.19568909, 0.19568909, 0.19568909, 0.20291032, 0.08585489,…## $ Poor <dbl> 0.032843150, 0.032843150, 0.032843150, 0.045383332, 0.00647…Actual vs. Predicted Health Rating
We can use our model to predict a person's perceived health rating given their age and whether they exercise
For each observation, the predicted perceived health rating is the category with the highest predicted probability
health_m_aug <- health_m_aug %>% mutate(pred_health = predict(health_m, type = "class"))Actual vs. Predicted Health Rating
health_m_aug %>% count(HealthGen, pred_health, .drop = FALSE) %>% pivot_wider(names_from = pred_health, values_from = n)## # A tibble: 5 x 6## HealthGen Excellent Vgood Good Fair Poor## <fct> <int> <int> <int> <int> <int>## 1 Excellent 0 550 223 0 0## 2 Vgood 0 1376 785 0 0## 3 Good 0 1255 1399 0 0## 4 Fair 0 300 642 0 0## 5 Poor 0 24 156 0 0Why do you think no observations were predicted to have a rating of "Excellent", "Fair", or "Poor"?
Why do you think no observations were predicted to have a rating of "Excellent", "Fair", or "Poor"?
Model selection
Comparing Nested Models
Suppose there are two models:
- Reduced Model includes predictors x1,…,xq
- Full Model includes predictors x1,…,xq,xq+1,…,xp
We want to test the hypotheses H0:βq+1=⋯=βp=0Ha: at least 1 βj is not0
To do so, we will use the Drop-in-Deviance test (very similar to logistic regression)
Add Education to the model?
We consider adding the participants'
Educationlevel to the model.- Education takes values
8thGrade,9-11thGrade,HighSchool,SomeCollege, andCollegeGrad
- Education takes values
Models we're testing:
- Reduced Model:
Age,PhysActive - Full Model:
Age,PhysActive,Education
- Reduced Model:
H0:β9−11thGrade=βHighSchool=βSomeCollege=βCollegeGrad=0Ha: at least one βj is not equal to 0
Add Education to the model?
H0:β9−11thGrade=βHighSchool=βSomeCollege=βCollegeGrad=0Ha: at least one βj is not equal to 0
model_red <- multinom(HealthGen ~ Age + PhysActive, data = nhanes_adult)model_full <- multinom(HealthGen ~ Age + PhysActive + Education, data = nhanes_adult)Add Education to the model?
anova(model_red, model_full, test = "Chisq") %>% kable(format = "markdown")| Model | Resid. df | Resid. Dev | Test | Df | LR stat. | Pr(Chi) |
|---|---|---|---|---|---|---|
| Age + PhysActive | 25848 | 16994.23 | NA | NA | NA | |
| Age + PhysActive + Education | 25832 | 16505.10 | 1 vs 2 | 16 | 489.1319 | 0 |
Add Education to the model?
anova(model_red, model_full, test = "Chisq") %>% kable(format = "markdown")| Model | Resid. df | Resid. Dev | Test | Df | LR stat. | Pr(Chi) |
|---|---|---|---|---|---|---|
| Age + PhysActive | 25848 | 16994.23 | NA | NA | NA | |
| Age + PhysActive + Education | 25832 | 16505.10 | 1 vs 2 | 16 | 489.1319 | 0 |
At least one coefficient associated with Education is non-zero. Therefore, we will include Education in the model.
Model with Education
| y.level | term | estimate | std.error | statistic | p.value | conf.low | conf.high |
|---|---|---|---|---|---|---|---|
| Vgood | (Intercept) | 0.582 | 0.301 | 1.930 | 0.054 | -0.009 | 1.173 |
| Vgood | Age | 0.001 | 0.003 | 0.419 | 0.675 | -0.004 | 0.006 |
| Vgood | PhysActiveYes | -0.264 | 0.099 | -2.681 | 0.007 | -0.457 | -0.071 |
| Vgood | Education9 - 11th Grade | 0.768 | 0.308 | 2.493 | 0.013 | 0.164 | 1.372 |
| Vgood | EducationHigh School | 0.701 | 0.280 | 2.509 | 0.012 | 0.153 | 1.249 |
| Vgood | EducationSome College | 0.788 | 0.271 | 2.901 | 0.004 | 0.256 | 1.320 |
| Vgood | EducationCollege Grad | 0.408 | 0.268 | 1.522 | 0.128 | -0.117 | 0.933 |
| Good | (Intercept) | 2.041 | 0.272 | 7.513 | 0.000 | 1.508 | 2.573 |
| Good | Age | -0.002 | 0.003 | -0.651 | 0.515 | -0.007 | 0.003 |
| Good | PhysActiveYes | -0.758 | 0.096 | -7.884 | 0.000 | -0.946 | -0.569 |
| Good | Education9 - 11th Grade | 0.360 | 0.275 | 1.310 | 0.190 | -0.179 | 0.899 |
| Good | EducationHigh School | 0.085 | 0.247 | 0.345 | 0.730 | -0.399 | 0.569 |
| Good | EducationSome College | -0.011 | 0.239 | -0.047 | 0.962 | -0.480 | 0.457 |
| Good | EducationCollege Grad | -0.891 | 0.236 | -3.767 | 0.000 | -1.354 | -0.427 |
| Fair | (Intercept) | 2.116 | 0.288 | 7.355 | 0.000 | 1.552 | 2.680 |
| Fair | Age | 0.000 | 0.003 | 0.107 | 0.914 | -0.006 | 0.006 |
| Fair | PhysActiveYes | -1.191 | 0.115 | -10.367 | 0.000 | -1.416 | -0.966 |
| Fair | Education9 - 11th Grade | -0.224 | 0.279 | -0.802 | 0.422 | -0.771 | 0.323 |
| Fair | EducationHigh School | -0.832 | 0.252 | -3.307 | 0.001 | -1.326 | -0.339 |
| Fair | EducationSome College | -1.343 | 0.246 | -5.462 | 0.000 | -1.825 | -0.861 |
| Fair | EducationCollege Grad | -2.509 | 0.253 | -9.913 | 0.000 | -3.005 | -2.013 |
| Poor | (Intercept) | -0.200 | 0.411 | -0.488 | 0.626 | -1.005 | 0.605 |
| Poor | Age | 0.018 | 0.005 | 3.527 | 0.000 | 0.008 | 0.028 |
| Poor | PhysActiveYes | -2.267 | 0.242 | -9.377 | 0.000 | -2.741 | -1.793 |
| Poor | Education9 - 11th Grade | -0.360 | 0.353 | -1.020 | 0.308 | -1.053 | 0.332 |
| Poor | EducationHigh School | -1.150 | 0.334 | -3.438 | 0.001 | -1.805 | -0.494 |
| Poor | EducationSome College | -1.073 | 0.316 | -3.399 | 0.001 | -1.692 | -0.454 |
| Poor | EducationCollege Grad | -2.322 | 0.366 | -6.342 | 0.000 | -3.039 | -1.604 |
Compare NHANES models using AIC
glance(model_red)$AIC## [1] 17018.23glance(model_full)$AIC## [1] 16561.1Compare NHANES models using AIC
glance(model_red)$AIC## [1] 17018.23glance(model_full)$AIC## [1] 16561.1Use the step() function to do model selection with AIC as the selection criteria
Checking conditions
Assumptions for multinomial logistic regression
We want to check the following assumptions for the multinomial logistic regression model:
Linearity: Is there a linear relationship between the log-odds and the predictor variables?
Randomness: Was the sample randomly selected? Or can we reasonably treat it as random?
Independence: There is no obvious relationship between observations
Checking linearity
Similar to logistic regression, we will check linearity by examining empirical logit plots between each level of the response and the quantitative predictor variables.
nhanes_adult <- nhanes_adult %>% mutate(Excellent = factor(if_else(HealthGen == "Excellent", "1", "0")), Vgood = factor(if_else(HealthGen == "Vgood", "1", "0")), Good = factor(if_else(HealthGen == "Good", "1", "0")), Fair = factor(if_else(HealthGen == "Fair", "1", "0")), Poor = factor(if_else(HealthGen == "Poor", "1", "0")) )Checking linearity
library(Stat2Data)par(mfrow = c(2,1))emplogitplot1(Excellent ~ Age, data = nhanes_adult, ngroups = 5, main = "Excellent vs. Age")emplogitplot1(Vgood ~ Age, data = nhanes_adult, ngroups = 5, main = "Vgood vs. Age")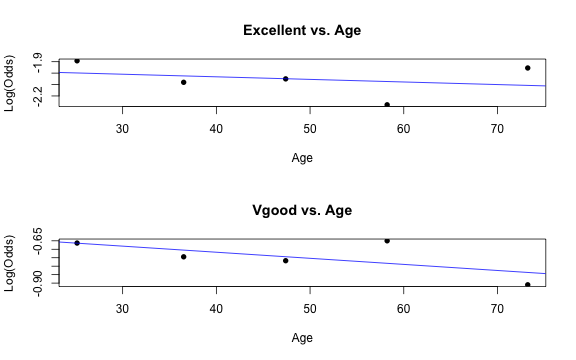
Checking linearity
par(mfrow = c(2,1))emplogitplot1(Good ~ Age, data = nhanes_adult, ngroups = 5, main = "Good vs. Age")emplogitplot1(Fair ~ Age, data = nhanes_adult, ngroups = 5, main = "Fair vs. Age")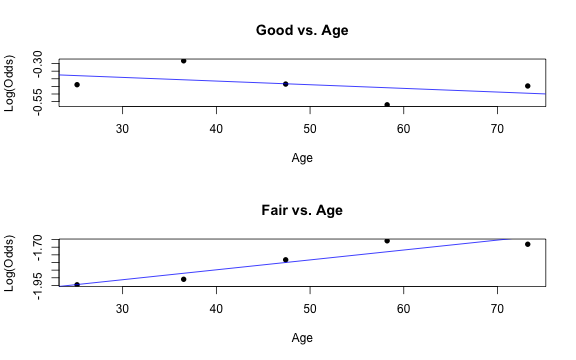
Checking linearity
emplogitplot1(Poor ~ Age, data = nhanes_adult, ngroups = 5, main = "Poor vs. Age")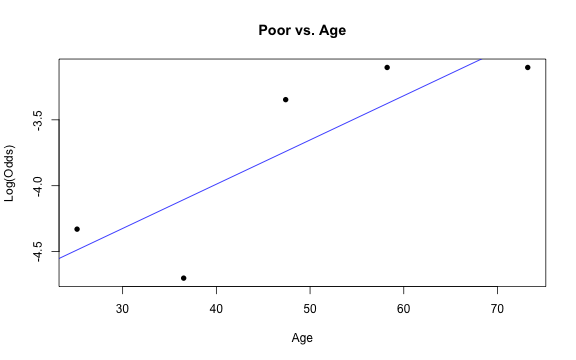
Checking linearity
emplogitplot1(Poor ~ Age, data = nhanes_adult, ngroups = 5, main = "Poor vs. Age")
✅ The linearity condition is satisfied. There is a linear relationship between the empirical logit and the quantitative predictor variable, Age.
Checking randomness
We can check the randomness condition based on the context of the data and how the observations were collected.
Was the sample randomly selected?
If the sample was not randomly selected, ask whether there is reason to believe the observations in the sample differ systematically from the population of interest.
Checking randomness
We can check the randomness condition based on the context of the data and how the observations were collected.
Was the sample randomly selected?
If the sample was not randomly selected, ask whether there is reason to believe the observations in the sample differ systematically from the population of interest.
✅ The randomness condition is satisfied. We do not have reason to believe that the participants in this study differ systematically from adults in the U.S..
Checking independence
We can check the independence condition based on the context of the data and how the observations were collected.
Independence is most often violated if the data were collected over time or there is a strong spatial relationship between the observations.
Checking independence
We can check the independence condition based on the context of the data and how the observations were collected.
Independence is most often violated if the data were collected over time or there is a strong spatial relationship between the observations.
✅ The independence condition is satisfied. It is reasonable to conclude that the participants' health and behavior characteristics are independent of one another.
Recap
Predictions
Model selection
Checking conditions